Energy efficient, MAPu uses a reductive glycine anabolic pathway to achieve carbon fixation and increase biomass production by 17%
“If the planet were a business, carbon dioxide would be its expense and carbon sequestration its income.”
At present, this “carbon account” is seriously empty.
Every year, human emissions of carbon dioxide far exceed the fixed capacity of nature, global warming, energy crisis is imminent.
C1 compounds such as formic acid and carbon dioxide are considered as key carbon sources for future bio-manufacturing due to their renewable and low-cost characteristics.
But for microbes to use them efficiently, the traditional natural carbon fixation pathway has become a bottleneck.
The Calvin cycle is the most common carbon sequestration pathway in the biological world, but its efficiency is limited.
Tobias Erb and colleagues at the Max Planck Institute for Terrestrial Microbiology have developed artificial carbon sequestration pathways, such as the CETCH and THETA cycles, that are more efficient than the natural Calvin cycle.
These pathways have been successful in vitro, but are only partially integrated into living organisms.
Another method of carbon sequestration they studied involves physicochemical methods, such as using renewable energy to electrochemically reduce CO₂ to formic acid, which is then converted into various products using microorganisms.
The researchers are currently developing a hybrid technology that first physically and chemically immobilizes CO₂ into formic acid and then further processes it through microorganisms.
Many bacteria naturally convert formic acid through the Calvin cycle, and the team recently tested a more energy efficient reducing glycinate pathway (rGlyP) than the Calvin cycle, which is the most efficient anabolic pathway for processing formic acid.
It was found that the engineered strains incorporating the rGlyP pathway produced 17% more biomass than the wild-type, and the yield was higher than any naturally-morphotrophic bacteria using the Calvin cycle.
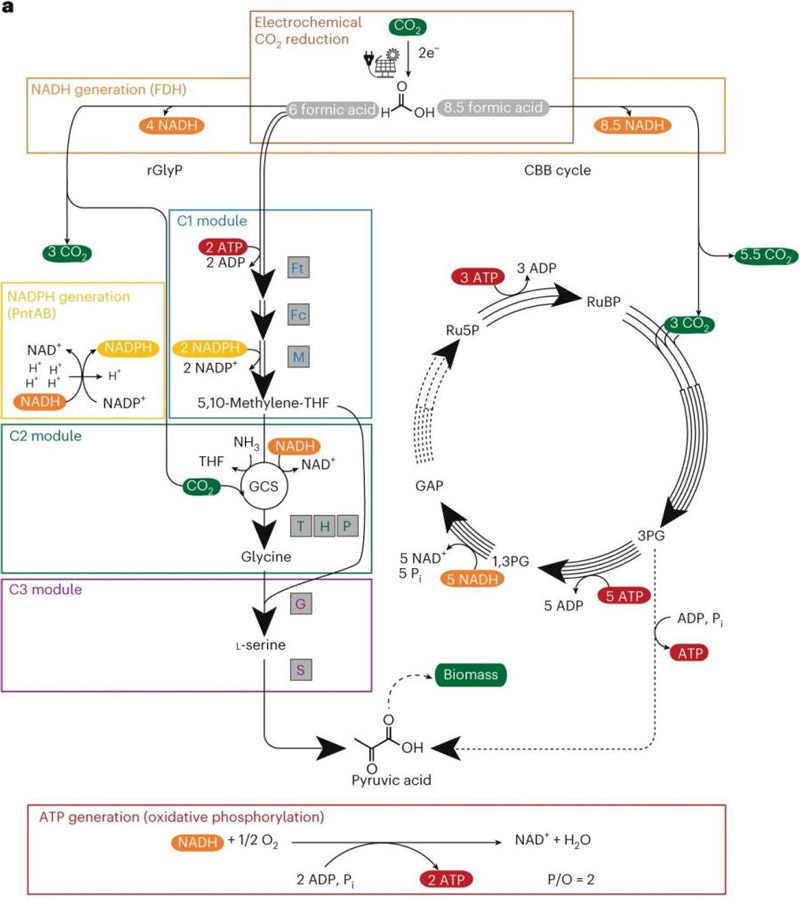
This suggests that anabolism has the potential to achieve sustainable biobased production. the study was published in the journal Nature under the title “One-carbon fixation via the synthetic reductive glycine pathway exceeds yield of the Calvin cycle. Microbiology.
For this study, the team used the Cupriavidus necator strain, which naturally uses the Calvin cycle to metabolize formic acid.
Back in 2020, one of the team’s collaborators, Nico Claassens from Wageningen University, had successfully introduced the reducing glycine pathway into Cupriavidus necator.
However, the resulting growth rate and biomass yield were lower than those of unmodified bacteria.
In the new study, the researchers integrated the complete reducing glycine pathway (rGlyP) into the bacterial genome and systematically optimized its efficiency.
The pathway consists of three key modules:
The C1 module activates formic acid through formate-tetrahydrofolate ligase (FtfL), and then generates methylene tetrahydrofolate cyclase (FchA) and dehydrogenase (MtdA).
The C2 module uses the glycine cracking system (GCS) to reverse combine with CO₂ to produce glycine, bypassing the energy-intensive Calvin cycle.
The C3 module converts glycine to pyruvate via serine hydroxymethyltransferase (GlyA) and deaminase (SdaA) – a series of reactions that ultimately produce pyruvate, which provides the core carbon backbone for cell growth.
The research team selected Cupriavidus necator H16, a commonly used strain in industry (naturally dependent on the Calvin cycle), as the chassis, and removed its polyhydroxybutyrate synthesis gene (ΔphaC1) by gene knockout technology to obtain an engineered strain H16δPHac1 with a clear metabolic background.
They isolated the C1 and C3 modules of rGlyP from the primordial plasmid system and integrated them into the bacterial genome in steps:
First, C1 module was randomly inserted into the genome using Tn5 transposon technique, and the fastest growing strains were screened by multiple rounds of formic acid culture medium passage.
On this basis, the same strategy was used again to integrate the C3 module, and finally the fully genome-integrated strain CRG6 was obtained.
To reduce metabolic shunt, the team further knocked out the glycine-oxidase gene (ΔdadA6), forcing the carbon flow to concentrate on rGlyP.
Proteomic analysis showed that genomic integration reduced the expression of RGLYp-associated proteins by 13%, and released cell resources were reallocated to growth-essential functions such as ribosome synthesis.
The biomass yield of the optimized CRG6 strain in formic acid medium reached 4.52 g CDW/mol, 17% higher than that of the wild type, and the carbon loss was reduced to 11%, showing significant metabolic efficiency advantages.
Through rational design and genome integration of anabolic pathways, this study has for the first time achieved a comprehensive surmount of natural carbon fixation systems in industrial microorganisms.
Its core value is not only reflected in the 17% increase in formic acid utilization efficiency, but also in the construction of a scalable “carbon-energy coupling” paradigm – the use of electrochemical reduction of formic acid generated by CO₂, through RGLYp-driven efficient absorption, closed-loop production of bioplastics (such as PHB), microbial proteins and C4-C6 liquid fuels. Reduce dependence on petroleum-based raw materials.
This technical framework shows significant cross-border potential.
The study was described as a “milestone in synthetic biology” (Sarwar & Lee, 2025), demonstrating that artificial metabolic networks can break through natural evolutionary constraints, and that future research needs to focus on “transforming theoretical advantages into industrial competitiveness” – as the review puts it: “making bacterial factories not only ‘smarter’ but also ‘more diligent.'”
Reference link:
- 1. Dronsella, B., Orsi, E., Schulz-Mirbach, H. et al. One-carbon fixation via the synthetic reductive glycine pathway exceeds yield of the Calvin cycle. Nat Microbiol (2025).
- 2. Sarwar, A., Lee, E.Y. Surpassing natural limits in one-carbon assimilation. Nat Microbiol (2025).